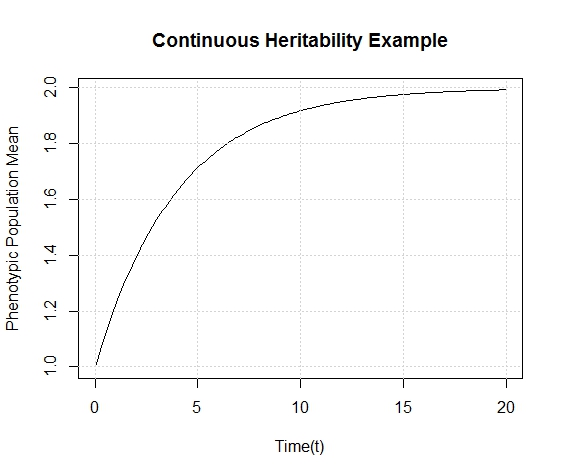
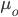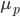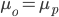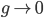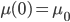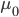Hops are what give beer a bitter, tangy, citrusy, earthy, fruity, floral, herbal, woody, piney, and/or a spicy aromas and flavors. Originally they were used as a preservative because of their natural antimicrobial powers, now they are a defining characteristic of beer. Now with the craft beer movement in full swing they are gaining lots of attention. Hoppy beers like IPAs and pale ales seem to be everyone’s favorites. These days, it seems every brewery is judged on its version of an IPA or pale ale. Everyone is falling behind the view that the hoppier a beer is, the better it must be. I’ve always disagreed with this and come to the realization that I only like a few hoppy beers, and I have to be in the mood for them. This leads me to ask the question, are there more people out there like me? Are hops craved seasonally?
To answer this question, I built an R web scraper to download all the top 100 beers and reviews/ratings/dates for each of those beers (code for that scraper will be posted on GitHub soon). Here is the first six rows of a data set with nearly 90,000 entries:
> head(data)
name date rating state
1 90 Minute IPA 2007-04-09 4.33 Illinois
2 90 Minute IPA 2007-04-07 4.80 California
3 90 Minute IPA 2007-04-02 4.85 Pennsylvania
4 90 Minute IPA 2005-07-03 4.47 Oregon
5 Adam From The Wood 2005-09-12 4.53 Ontario (Canada)
6 AleSmith IPA 2013-09-30 4.20 Wisconsin
review
1 redoing this as my review was unceremoniously...
2 Received in a trade from geexploitation. An excellent...
3 This was really an exceptional beer from start to finish...
4 This IPA pours a beautiful golden color, it leaves...
5 Poured with a medium amount of heat behind it the...
6 The liquid portion is a hazy marmalade orange color...
style abv brewery
1 American Double / Imperial IPA 9.00 Dogfish Head Brewery
2 American Double / Imperial IPA 9.00 Dogfish Head Brewery
3 American Double / Imperial IPA 9.00 Dogfish Head Brewery
4 American Double / Imperial IPA 9.00 Dogfish Head Brewery
5 Old Ale 12.00 Hair of the Dog Brewing Company / Brewery and Tasting Room
6 American IPA 7.25 AleSmith Brewing Company
link
1 http://beeradvocate.com/beer/profile/10099/2093/
2 http://beeradvocate.com/beer/profile/10099/2093/
3 http://beeradvocate.com/beer/profile/10099/2093/
4 http://beeradvocate.com/beer/profile/10099/2093/
5 http://beeradvocate.com/beer/profile/173/20767/
6 http://beeradvocate.com/beer/profile/396/3916/
Yup, the first four entries are for the world famous Dogfish Head 90 minute IPA, a classic representation of this style. What styles are there and what are we including in ‘Hoppy Beers’? The styles I chose are “American Double”, “Imperial IPA”, “American IPA”, and “American Pale Ale”. There are many styles that are hoppy, and you might ask, “What about an English pale ale or an Extra Special Bitter?”, and the reason they weren’t included is that those styles didn’t appear in the top 100 at the time the data was retrieved.
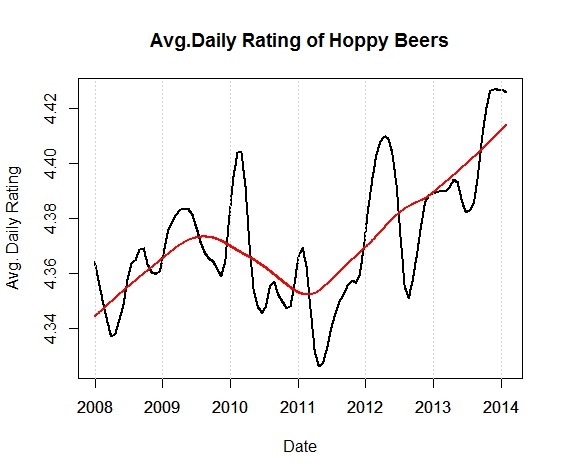
Looking at the exact point plot of ratings over time is meaningless, so let’s look at a smoothed representation of these beers.
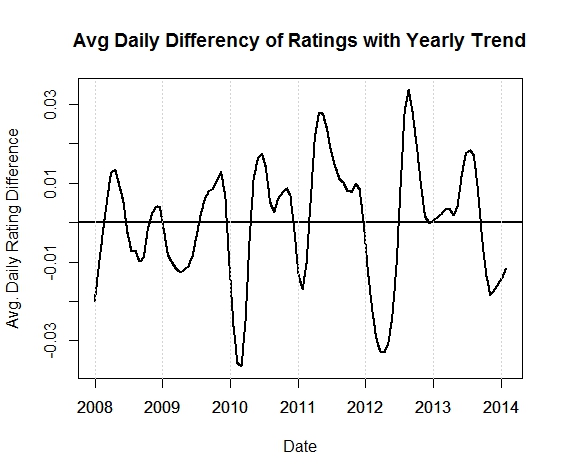
Definitely a trend upwards. This could be the hoppy craft beer culture at work here. What about seasonality? I added an over-smoothed line (in red) to the chart to see the yearly trend. To look at the seasonality within years, let’s subtract the two lines and remove the yearly trend.
Hmmmm, maybe a possible dip every year in the late winter/spring and a peak in the fall? Interesting. Let’s verify seasonality by finding the peak of the Fourier spectrum of this signal. This will tell us the dominant frequencies in the above time series, which turns out to be around f=0.003006012 days. To find the period of this frequency we invert it to get:
T = 332.67 days.
ALMOST ONE YEAR.
I bet with more data, and controlling for quantity, we could narrow this down to exactly one year. In the future I’m going to do some resampling to figure out the error in the period and hopefully 365.25 will fall within the error bounds. For now, I think I’m going to have a beer.
Cheers.
R code for analysis:
library(TSA) # Time series analysis package
##----Plot Ratings of Hoppy Beers----
hoppy_beerstyles = c("American Double / Imperial IPA","American IPA","American Pale Ale (APA)")
date.seq = seq(from=as.Date("2008-01-01"),to=range(data$date)[2],by=1)
hoppy_ratings_avg = sapply(date.seq,function(x){
mean(data$rating[data$style%in%hoppy_beerstyles & data$date==x],na.rm=TRUE)
})
date.seq = date.seq[!is.na(hoppy_ratings_avg)]
hoppy_ratings_avg = hoppy_ratings_avg[!is.na(hoppy_ratings_avg)]
hoppy_ratings_smooth = lowess(date.seq,hoppy_ratings_avg,f=0.08)$y
hoppy_over_smooth = lowess(date.seq,hoppy_ratings_avg,f=0.4)$y
grid_lines=seq(as.Date("2008-01-01"),as.Date("2014-01-01"), by="+6 month")
plot(date.seq,hoppy_ratings_smooth,main="Avg.Daily Rating of Hoppy Beers",type="l",
xlab="Date",ylab="Avg. Daily Rating",lwd=2)
abline(v=axis.Date(1, x=grid_lines),col = "lightgray", lty = "dotted", lwd = par("lwd"))
lines(date.seq,hoppy_over_smooth,col="red",lwd=2)
##----remove hoppy trend/find periodicity-----
hoppy_no_trend = hoppy_over_smooth - hoppy_ratings_smooth
plot(date.seq,hoppy_no_trend,main="Avg Daily Differency of Ratings with Yearly Trend",type="l",
xlab="Date",ylab="Avg. Daily Rating Difference",lwd=2)
abline(h=0,lwd=2)
abline(v=axis.Date(1, x=grid_lines),col = "lightgray", lty = "dotted", lwd = par("lwd"))
# periodogram(hoppy_no_trend)
hoppy_spec = spectrum(hoppy_no_trend, method = "ar")
hoppy_spec_max = hoppy_spec$freq[which.max(hoppy_spec$spec)]